Dr. Guanjue Xiang

Senior Data Scientist | Computational Biology

Senior Data Scientist | Computational Biology
I am a researcher with expertise in computational biology, machine learning, and genomics. My research focuses on developing innovative computational methods for analyzing complex biological data, particularly in epigenomics and gene regulation.
My recent work includes developing machine learning systems for enhancer-promoter pair prediction and optimizing Anti-Sense Oligonucleotides design. I've published several high-impact papers in journals such as Nucleic Acids Research, Genome Research, and Nature Communications, with significant contributions to computational methods. My expertise spans from traditional statistical approaches to cutting-edge deep learning applications in genomics.
Cambridge, MA | 2022 - Present
Boston, MA | 2020 - 2022
Mentors: Dr. X. Shirley Liu and Dr. Jun Liu
University Park, PA | 2015 - 2020
Mentors: Dr. Yu Zhang and Dr. Ross C. Hardison
Minor in Applied Statistics
Penn State University, 2015-2020
Thesis: Computational Tools for Normalization, Integration, and Gene Regulatory Inference of Multi-Dimensional Epigenomic and Transcriptomic Data
Xiang, G., et al. (2024). "JMnorm: a novel joint multi-feature normalization method for integrative and comparative epigenomics." Nucleic Acids Research 52.2: e11-e11.

Figure: Overview of the JMnorm method for joint multi-feature normalization
Xiang, G., et al. (2024). "Interspecies regulatory landscapes and elements revealed by novel joint systematic integration of human and mouse blood cell epigenomes." Genome Research: gr-277950.
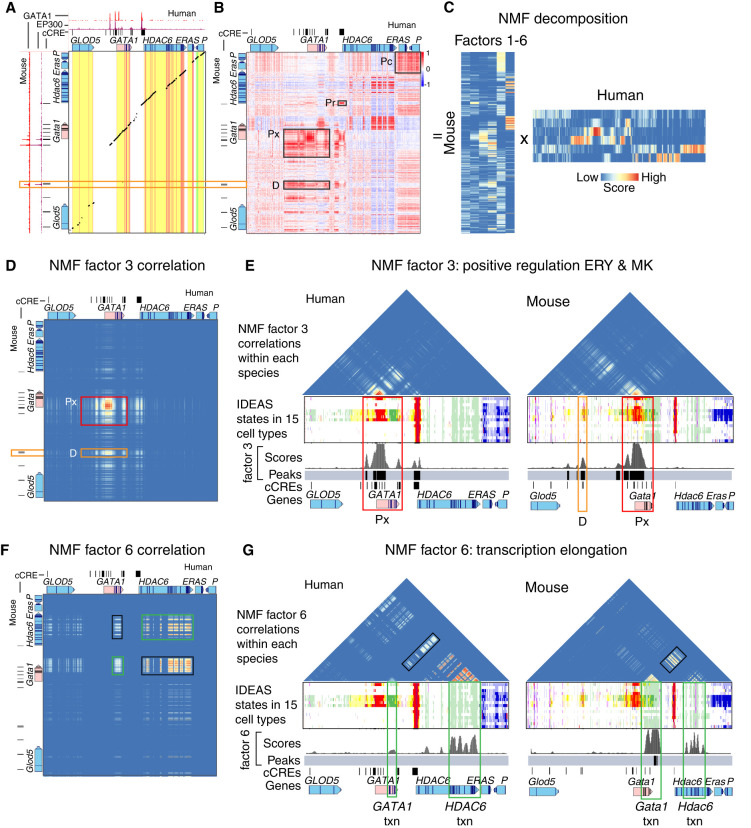
Figure: Interspecies regulatory landscapes and elements in human and mouse blood cells
Zhang, Y., Xiang, G., et al. (2023). "MetaTiME integrates single-cell gene expression to characterize the meta-components of the tumor immune microenvironment." Nature Communications 14.1: 2634.
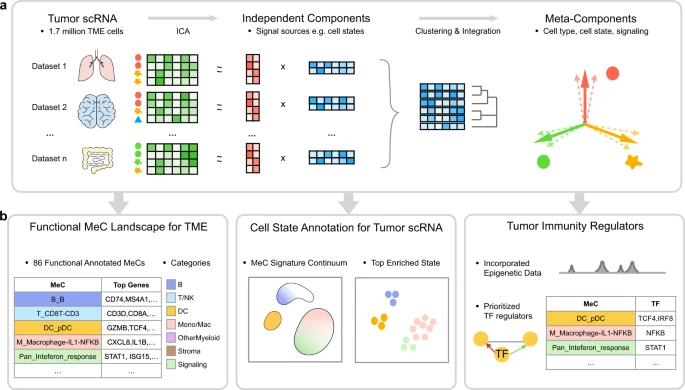
Figure: Overview of MetaTiME
Luan, J., Xiang, G., et al. (2021). "Distinct properties and functions of CTCF revealed by a rapidly inducible degron system." Cell Report 34.8: 108783.

Figure: Highly variable CTCF persistence on chromatin following auxin-mediated degradation
Xiang, G., et al. (2023). "Snapshot: a package for clustering and visualizing epigenetic history during cell differentiation." BMC Bioinformatics 24.1: 102.
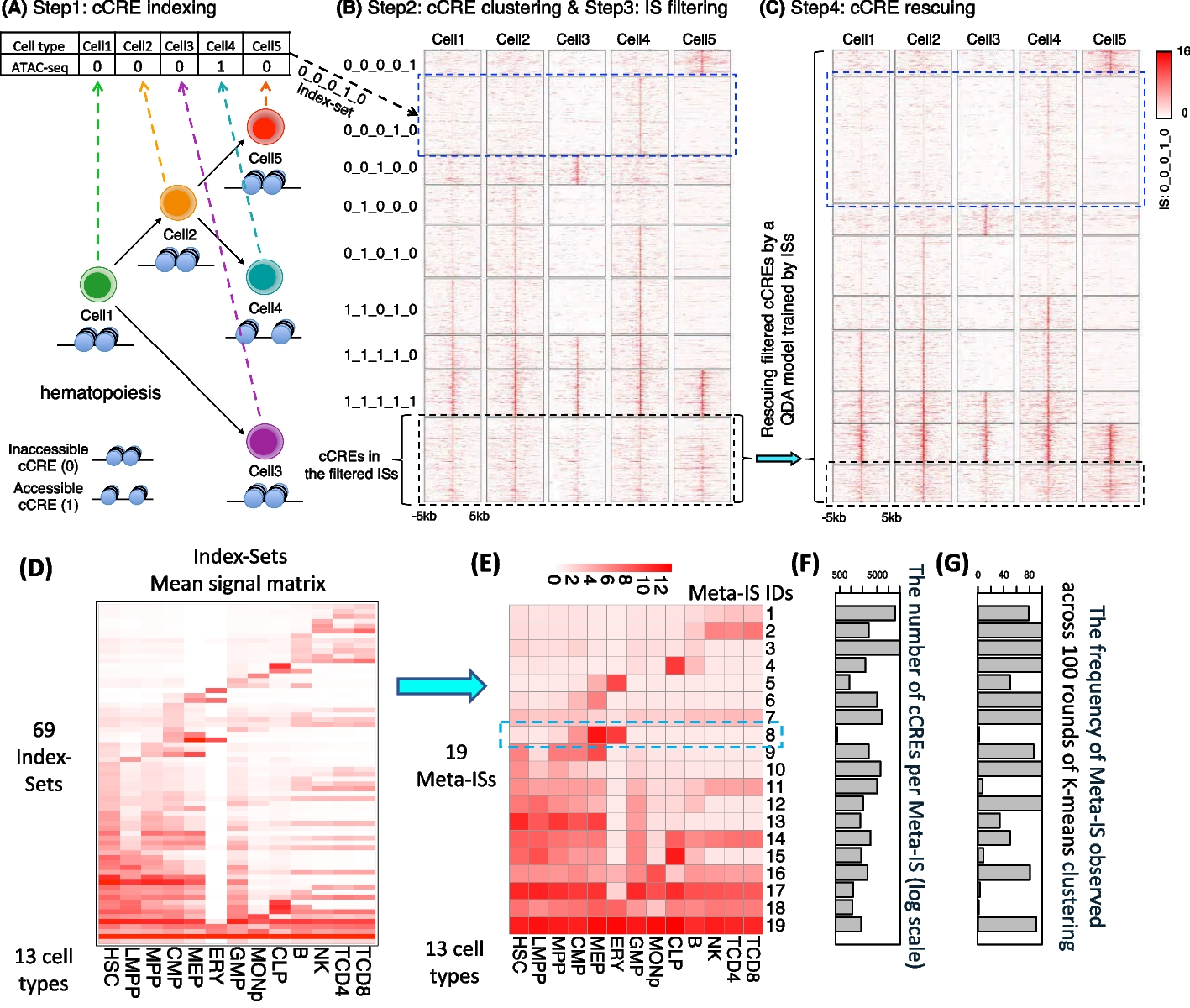
Figure: Overview of Snapshot package workflow
Xiang, G., et al. (2021). "S3V2-IDEAS: a package for normalizing, denoising and integrating epigenomic datasets across different cell types." Bioinformatics 37.13: 1847-1849.

Figure: Overview of S3V2-IDEAS package workflow
Xiang, G., Zhang, Y.*, Hardison, R.* (2020). "S3norm: simultaneous normalization of sequencing depth and signal-to-noise ratio in epigenomic data." Nucleic Acids Research 48.8: e43-e43.

Figure: Overview of S3norm method
Xiang, G., Keller, C., et al. (2020). "An integrative view of the regulatory and transcriptional landscapes in mouse hematopoiesis." Genome Research 30.3: 472-484.
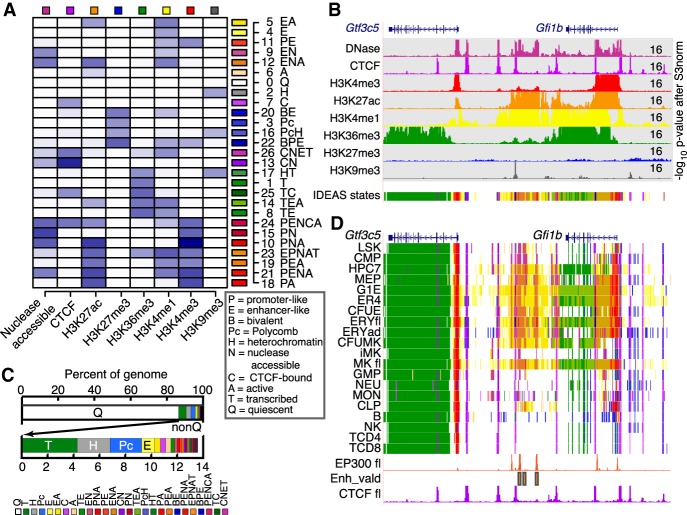
Figure: Overview of mouse hematopoiesis regulatory landscape
Zeng, Q., Xiang, G., et al. "JOnTADS: a unified caller for TADs and stripes in Hi-C data." (Manuscript in submission).